Getting started with moultmcmc
getting-started.RmdTo install moultmcmc please follow the instructions in
the README
file.
For demonstration purposes we use the sanderlings
dataset from Underhill and Zucchini
(1988). It contains observations of sampling dates
Day and moult indices MIndex for 164 adult
Sanderlings (Calidris alba) trapped on 11 days in the South
Africa in the austral summer of 1978/79. The sample contains 85
pre-moult individuals, 66 in active moult and 13 post-moult individuals.
The data are augmented with a column describing categorical moult status
MCat, so we can demonstrate fitting Type 1 models when data
are coded categorically.
data("sanderlings")
sanderlings$MCat <- case_when(sanderlings$MIndex == 0 ~ 1,
sanderlings$MIndex == 1 ~3,
TRUE ~ 2)
#plot the data
ggplot(sanderlings, aes(y=MIndex, x=Day, col = factor(MCat))) + geom_point() +theme_classic()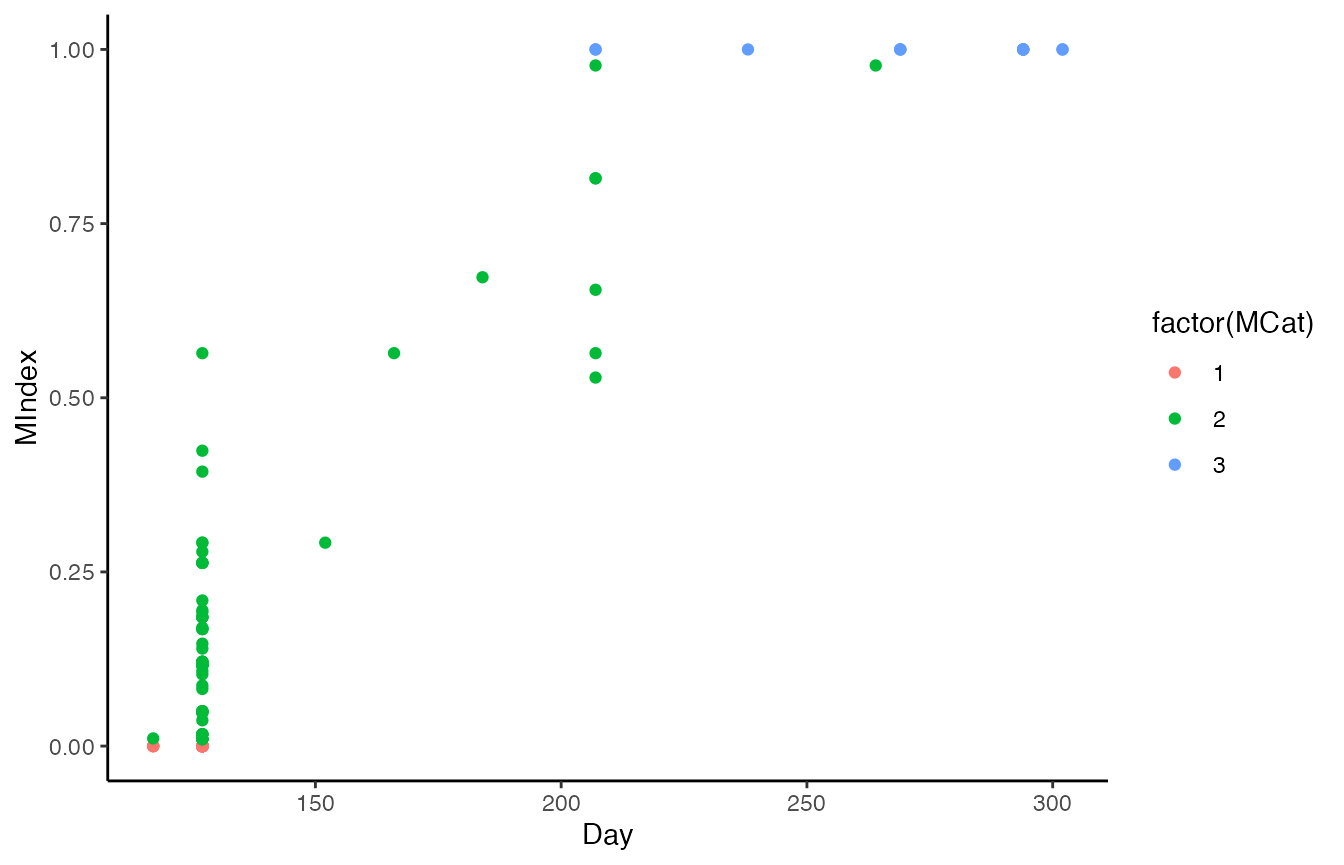
The central function for model fitting is the
moultmcmc() function. Model variants of the standard
Underhill-Zucchini models are specified using the type
argument to designate the relevant input data type. Additionally the
user has to designate the input data columns containing moult records,
and dates, respectively, as well as linear predictor structures for the
main moult parameters - the start date, duration and the standard
deviation of the start date. To fit an intercept-only type 1 model we
therefore use the following specification:
mcmc1 = moultmcmc(moult_column = "MCat",
date_column = "Day",
start_formula = ~1,
duration_formula = ~1,
sigma_formula = ~1,
data = sanderlings,
type=1,
init = "auto",
chains = 2,
refresh=1000)
#>
#> SAMPLING FOR MODEL 'uz1_linpred' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 5.2e-05 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.52 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 0.291 seconds (Warm-up)
#> Chain 1: 0.148 seconds (Sampling)
#> Chain 1: 0.439 seconds (Total)
#> Chain 1:
#>
#> SAMPLING FOR MODEL 'uz1_linpred' NOW (CHAIN 2).
#> Chain 2:
#> Chain 2: Gradient evaluation took 1.9e-05 seconds
#> Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.19 seconds.
#> Chain 2: Adjust your expectations accordingly!
#> Chain 2:
#> Chain 2:
#> Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 2:
#> Chain 2: Elapsed Time: 0.285 seconds (Warm-up)
#> Chain 2: 0.14 seconds (Sampling)
#> Chain 2: 0.425 seconds (Total)
#> Chain 2:Most MCMC parameters, such as the number of chains (here 2),
iterations, or cores are set in the same way as for
rstan::sampling, however, for initial values
moultmcmc defaults to an additional option
"auto" which will provide initial values for the MCMC
chains based on the input data. Custom initial values can be specified
instead as detailed in the documentation of
rstan::sampling.
Once the MCMC has completed, a summary table of parameter estimates can be displayed with
summary_table(mcmc1)
#> # A tibble: 5 × 11
#> parameter estimate se_mean sd lci uci n_eff Rhat prob method
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 mean_(Interc… 133. 0.0980 3.46 128. 141. 1244. 1.00 0.95 MCMC
#> 2 duration_(In… 97.9 0.294 10.6 78.0 119. 1303. 1.00 0.95 MCMC
#> 3 log_sd_(Inte… 3.32 0.00651 0.233 2.86 3.77 1286. 1.00 0.95 MCMC
#> 4 sd_(Intercep… 28.6 0.190 6.70 17.5 43.4 1242. 1.00 0.95 MCMC
#> 5 lp__ -104. 0.0433 1.24 -107. -103. 821. 1.00 0.95 MCMC
#> # ℹ 1 more variable: type <chr>and standard model assessment can be done as for any posterior sample
from Stan, by directly accessing the stanfit slot of the
returned S3 object. For example we can look at the traceplots for the
model using rstan::stan_trace
stan_trace(mcmc1$stanfit)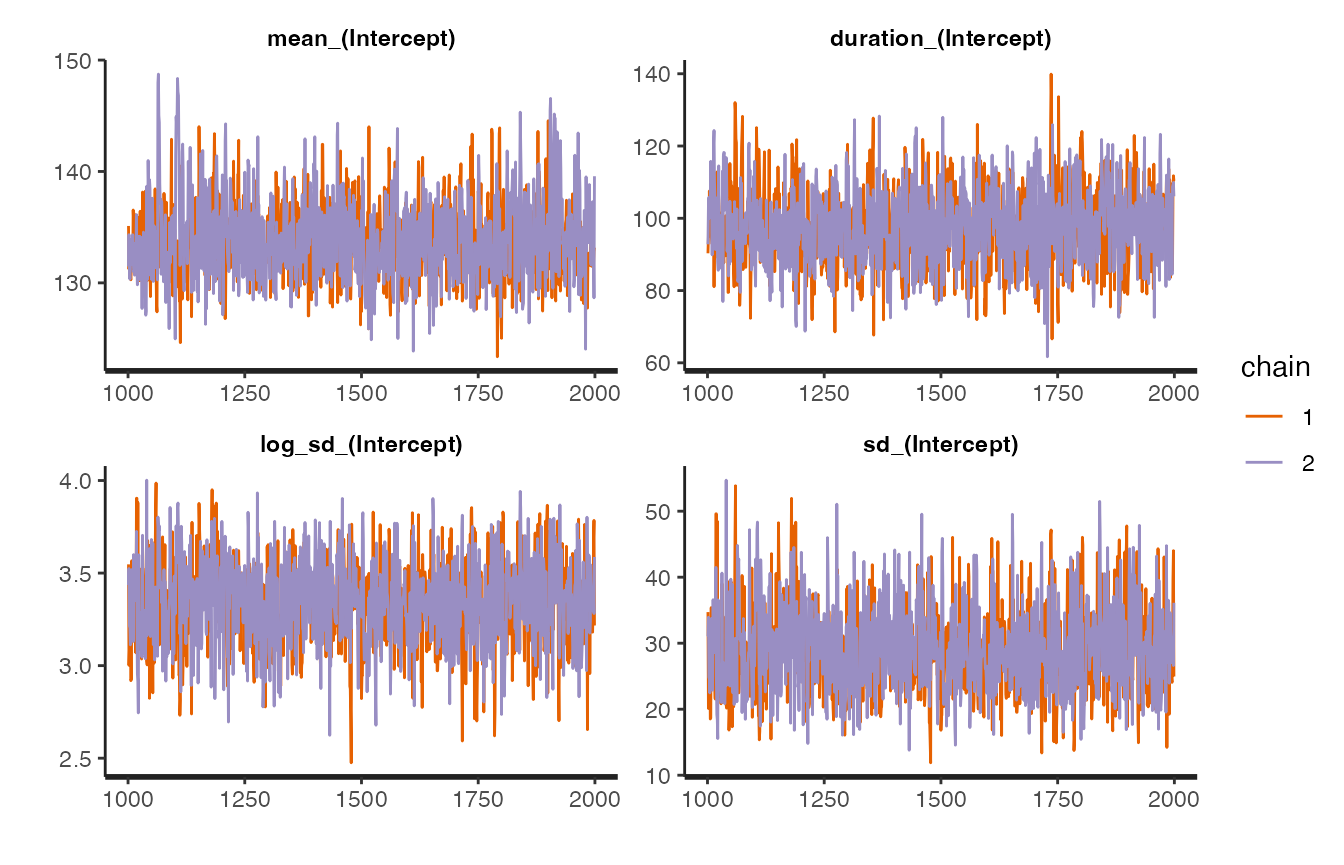 We can also plot the model fit against the data using the
We can also plot the model fit against the data using the
moult_plot function
moult_plot(mcmc1)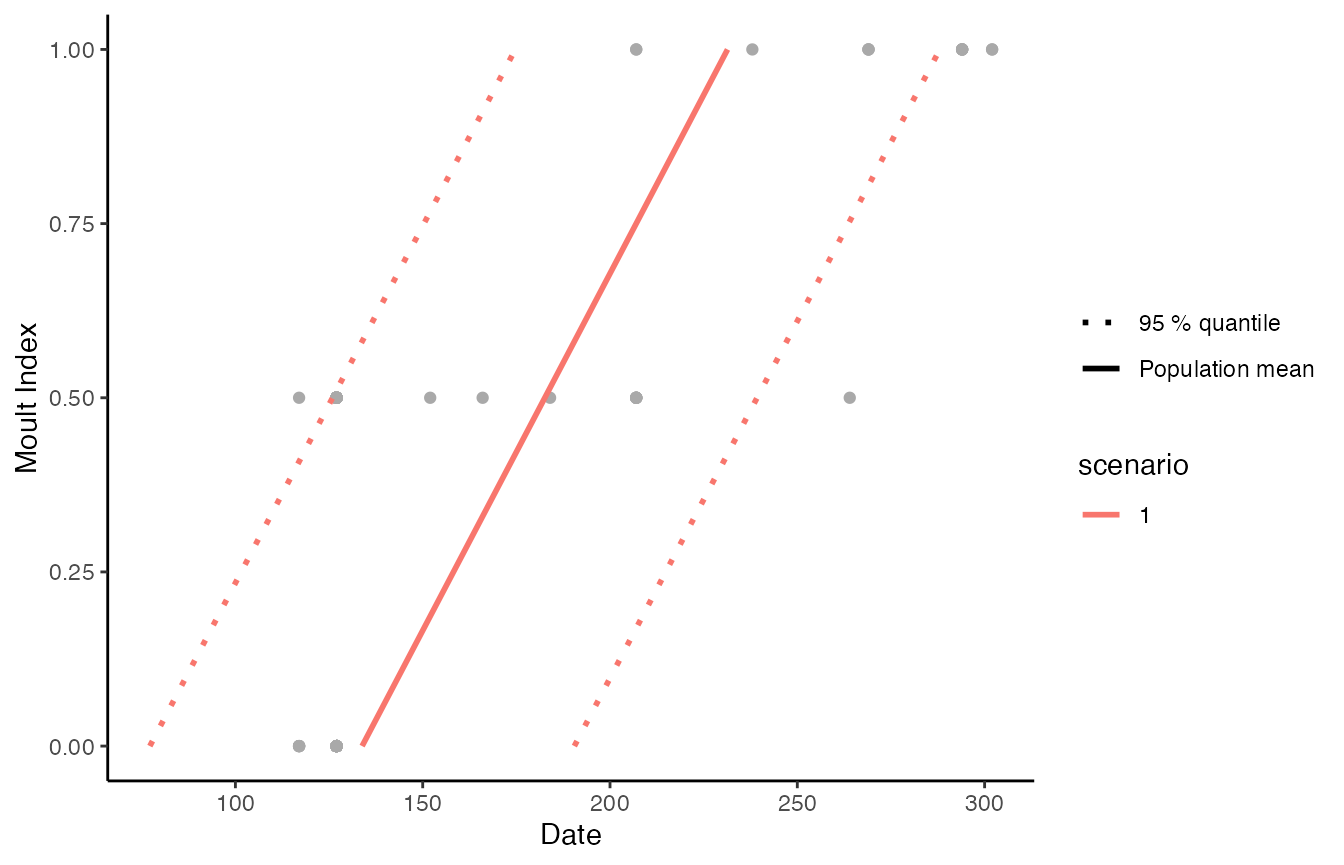
For comparison we can fit the same model using maximum likelihood
estimation via the moult package (Erni et al. 2013) and compare parameter
estimates visually with the compare_plot function.
ml1 = moult(MIndex ~ Day,data = sanderlings, type = 1)
compare_plot(ml1, mcmc1)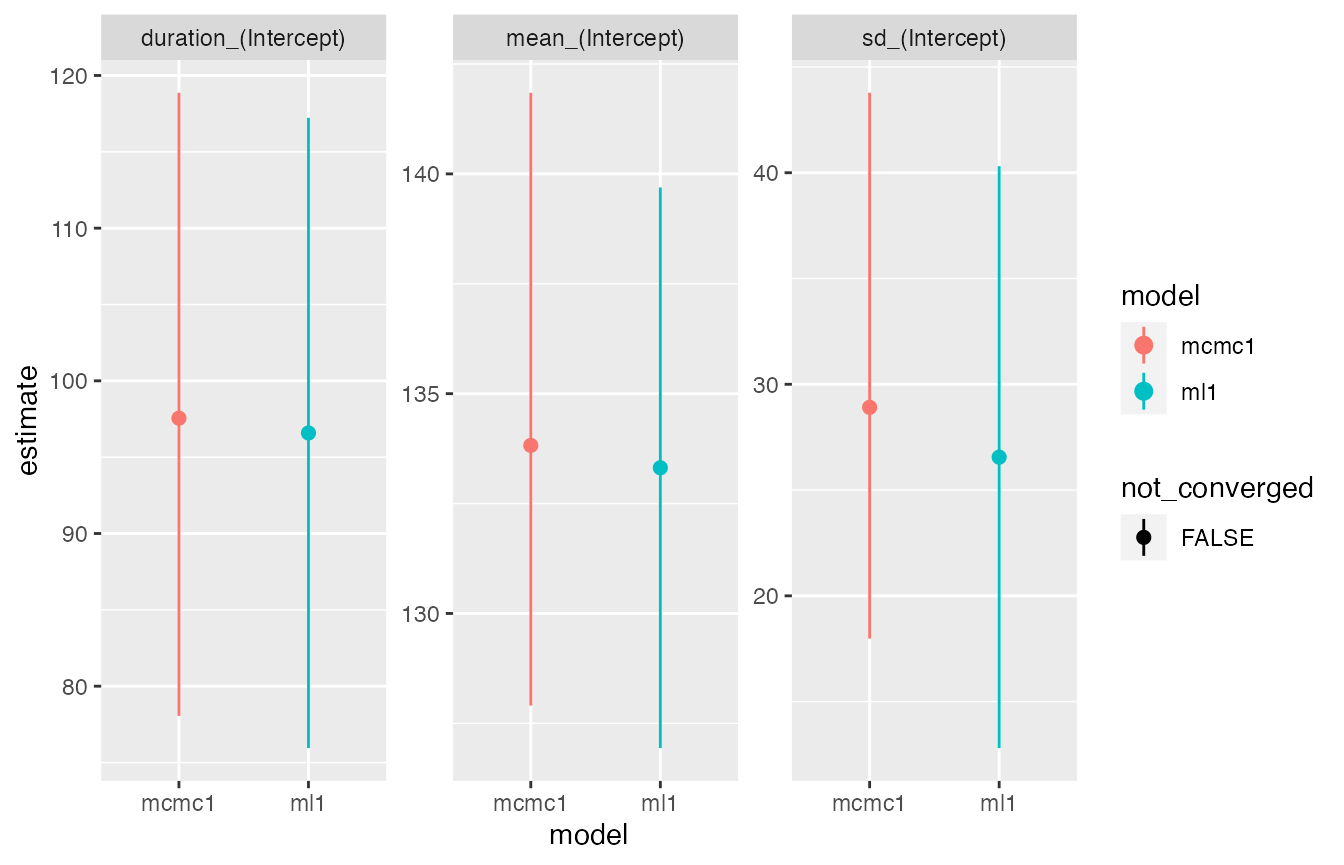
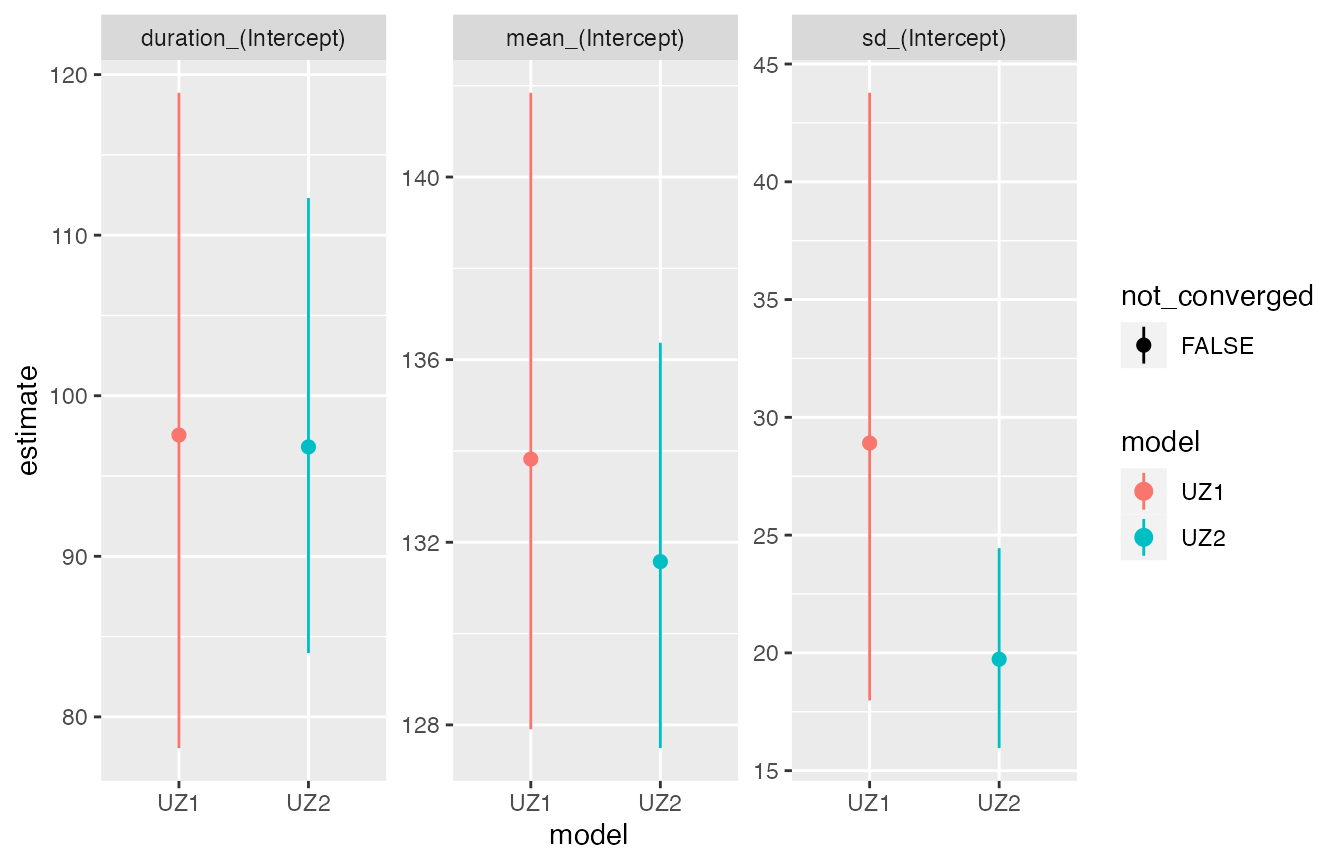
The comparison plot shows very similar estimates for both methods.
Similarly we can fit the corresponding Type 2 model, that makes use
of the full information contained in the moult indices, by setting
type=2. All moultmcmc models default to an
intercept only model, so we do not need to explicitly specify the
individual linear predictors here.
#fit using MCMC
mcmc2 = moultmcmc(moult_column = "MIndex",
date_column = "Day",
type=2,
data = sanderlings,
chains = 2,
refresh = 1000)
#>
#> SAMPLING FOR MODEL 'uz2_linpred' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 3e-05 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.3 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 0.237 seconds (Warm-up)
#> Chain 1: 0.099 seconds (Sampling)
#> Chain 1: 0.336 seconds (Total)
#> Chain 1:
#>
#> SAMPLING FOR MODEL 'uz2_linpred' NOW (CHAIN 2).
#> Chain 2:
#> Chain 2: Gradient evaluation took 1.7e-05 seconds
#> Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.17 seconds.
#> Chain 2: Adjust your expectations accordingly!
#> Chain 2:
#> Chain 2:
#> Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 2:
#> Chain 2: Elapsed Time: 0.207 seconds (Warm-up)
#> Chain 2: 0.127 seconds (Sampling)
#> Chain 2: 0.334 seconds (Total)
#> Chain 2:
#fit using ML
ml2 = moult(MIndex ~ Day,data = sanderlings, type = 2)
#compare parameter estimates
compare_plot(ml2, mcmc2)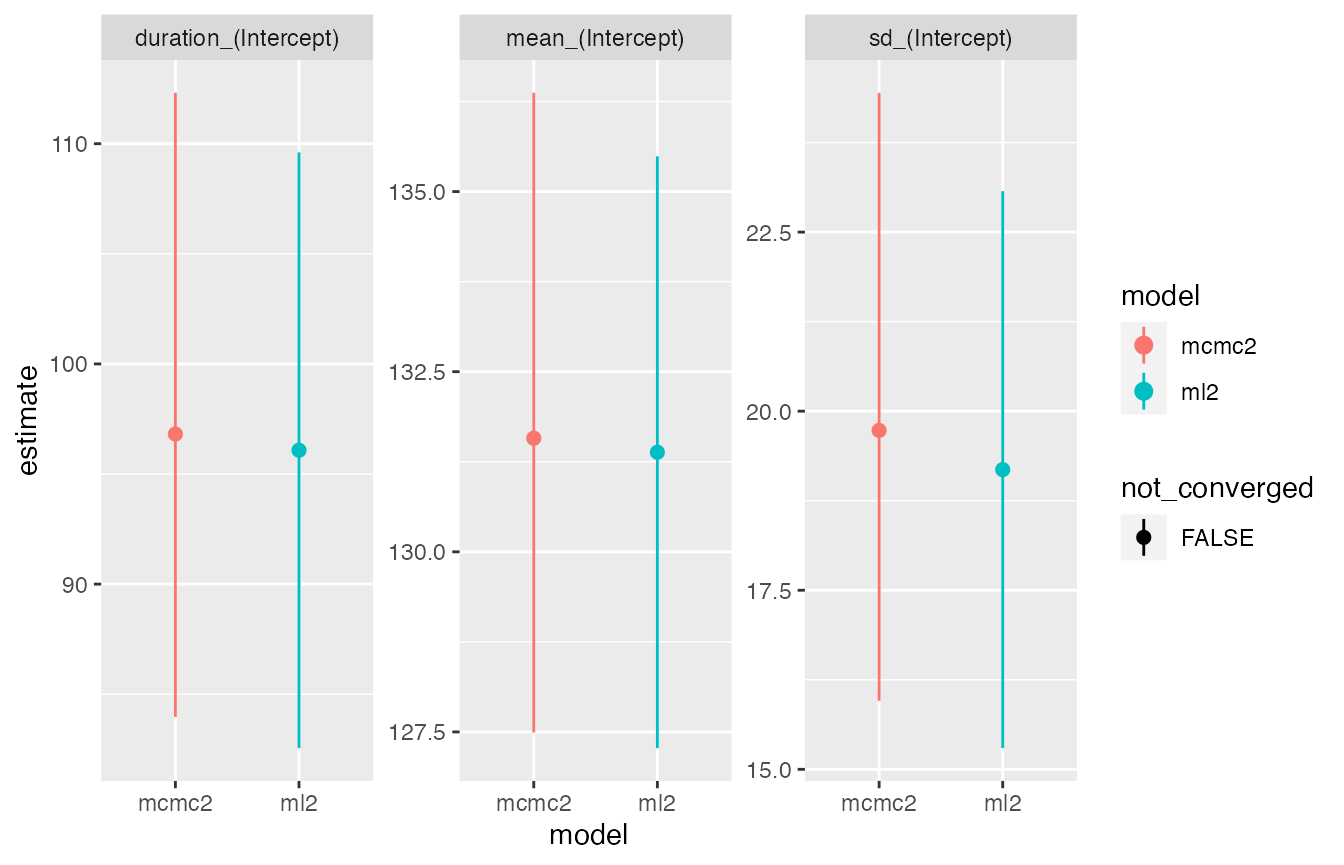
As before the comparison plot shows very similar estimates for both
methods. Perhaps more interesting is a comparison of the two model
types, which illustrates the gain in precision achieved by exploiting
the information contained in the moult indices. Note that models can be
given custom names for visualisation purposes using the
names argument to compare_plot
compare_plot(mcmc1, mcmc2, names = c('UZ1','UZ2'))