14 Nov 2016
Philipp Boersch-Supan
Seabirds
Tweet this!
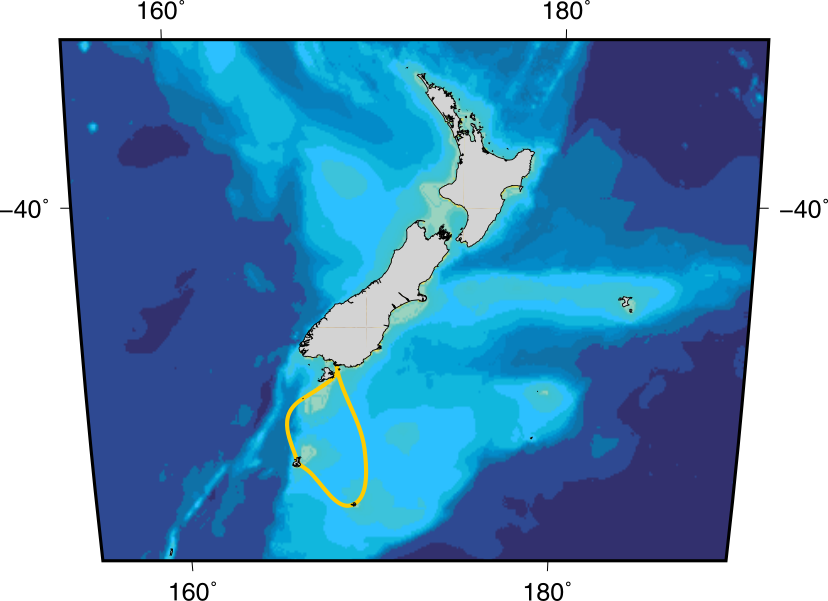
I am beyond thrilled to have been awarded a travel scholarship by the Enderby Trust, to go on a trip to the sub-Antarctic islands of New Zealand. In mid-December I will be visiting Tini Heke (Snares Island), Motu Maha (Auckland Islands), and Motu Ihupuku (Campbell Island), and – weather allowing – some of the seabird colonies on them. I am very excited to get this chance to see the species I am modelling in their natural habitats.
I will not be able to update the blog during the expedition, as there is no regular internet connection on the vessel. The vessel position and cccasional updates from the expedition leader can be found at http://www.heritage-expeditions.com/captains-blog/.
07 Nov 2016
Philipp Boersch-Supan
R Software Data mining
Tweet this!



I am very happy to announce a new software paper and R package published today in The Journal of Open Source Software and on CRAN. rucrdtw is a wrapper for the UCR Suite, a C++ library for very fast time series similarity searches under dynamic time warping (DTW).
Dynamic Time Warping (DTW) methods provide algorithms to optimally map a given time series onto all or part of another time series (Berndt and Clifford 1994). The remaining cumulative distance between the series after the alignement is a useful distance metric in time series data mining applications for tasks such as classification, clustering, and anomaly detection.
Calculating a DTW alignment is computationally relatively expensive, and as a consequence DTW is often a bottleneck in time series data mining applications. The UCR Suite (Rakthanmanon et al. 2012) provides a highly optimized algorithm for best-match subsequence searches that avoids unnecessary distance computations and thereby enables fast DTW and Euclidean Distance queries even in data sets containing trillions of observations.
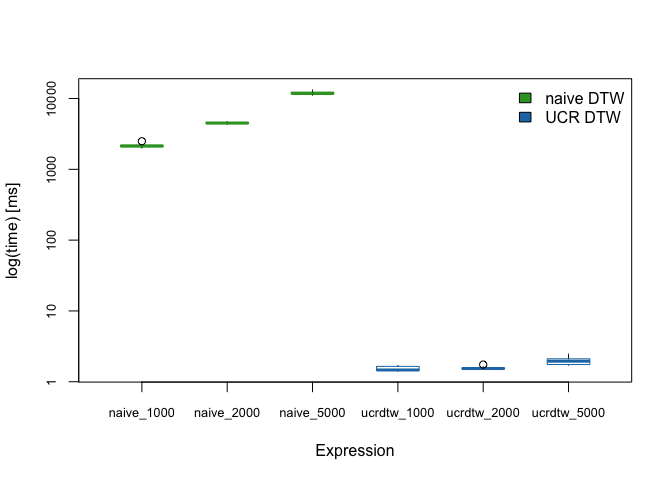
A broad suite of DTW algorithms is implemented in R in the dtw package (Giorgino 2009). The rucrdtw R package provides complementary functionality for fast similarity searches by providing R bindings for the UCR Suite via Rcpp (Eddelbuettel and Francois 2011). In addition to queries and data stored in text files, rucrdtw also implements methods for queries and/or data that are held in memory as R objects, as well as a method to do fast similarity searches against reference libraries of time series.
References
Berndt, Donald J, and James Clifford. 1994. “Using Dynamic Time Warping to Find Patterns in Time Series.” In KDD Workshop, 10:359–70. 16. AAAI. http://www.aaai.org/Library/Workshops/1994/ws94-03-031.php.
Eddelbuettel, Dirk, and Romain Francois. 2011. “Rcpp: Seamless R and C++ Integration.” Journal of Statistical Software 40 (1): 1–18. doi:10.18637/jss.v040.i08.
Giorgino, Toni. 2009. “Computing and Visualizing Dynamic Time Warping Alignments in R: The Dtw Package.” Journal of Statistical Software 31 (7): 1–24. doi:10.18637/jss.v031.i07.
Rakthanmanon, Thanawin, Bilson Campana, Abdullah Mueen, Gustavo Batista, Brandon Westover, Qiang Zhu, Jesin Zakaria, and Eamonn Keogh. 2012. “Searching and Mining Trillions of Time Series Subsequences Under Dynamic Time Warping.” In Proceedings of the 18th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 262–70. ACM. doi:10.1145/2339530.2339576.
15 Sep 2016
Philipp Boersch-Supan
Bayes R Software
Tweet this!

We are very excited to announce the initial CRAN release of our R package for Bayesian inference in differential equation models. At this point we consider the package API stable, and we will strive to preserve backwards compatibility in future releases. Also included in the CRAN release are three vignettes covering example applications for ODEs and DDEs.
The CRAN version of the package can be found at https://cran.r-project.org/package=deBInfer and the package can be installed in R with thecommand install.packages("deBInfer").
We continue to work on the package, and a development version is available at https://github.com/pboesu/debinfer it can be installed directly from github using the devtools package:
install.packages("devtools")
devtools::install_github("pboesu/debinfer")
HTML help files for the development version, and the most recent versions of the vignettes are available at http://pboesu.github.io/debinfer
Background on the methods implemented in deBInfer can be found in our paper deBInfer: Bayesian inference for dynamical models of biological systems in R
04 Jul 2016
Philipp Boersch-Supan
Bayes Albatrosses
Tweet this!
We have been busy spreading the word about our new R package for implementing Bayesian inference for differential equation models. Our poster from the 2016 meeting of the International Society of Bayesian Analysis and slides from Leah’s talk at the International Statistical Ecology Conference are now available on figshare. For more detail have a look at the arXiv preprint, or better yet, download the package from github, and try it.
At ISEC we also presented a poster featuring some early results on our work on albatross movement models, and their identifiability when using data from different generations of biologgers.
�
03 May 2016
Philipp Boersch-Supan
Bayes
Tweet this!
New preprint with Leah Johnson on Bayesian inference methods for differential equation models of biological systems: deBInfer: Bayesian inference for dynamical models of biological systems in R. Our paper describes an R package for implementing these kinds of analyses. The package itself is still pretty rough around the edges, but the development version is available from github, and includes a vignette featuring a worked example.


